9 Trends That Will Carry Genomics Into 2020 (And Beyond)

By Alex Vadas and Brian Baranick, L.E.K. Consulting
Remarkable advances in genomics technologies bring the promise of extraordinary changes in healthcare — and some of those changes are arriving now. What’s unfolding are nine trends that we think will shape the life science markets in this accelerating genomics revolution. First, some background on how we got here.
The field of genomics has surpassed expectations over the past three decades, due to massive changes in technology that allowed researchers to interrogate larger pieces of the human genome. The modern era of genomics arguably began in the mid-1980s with the development of the polymerase chain reaction (PCR) technique, which enabled researchers to characterize the genome at the candidate gene level. In the early 1990s, scientists leveraged semiconductor manufacturing techniques to develop microarrays that enabled large-scale genotyping and gene expression profiling studies. In 2003, at a cost of around $3 billion, the first human genome was sequenced. Since then, next generation sequencing (NGS) has dramatically decreased in cost, recently breaking the barrier of $1,000 per human genome.
These enabling technologies have not only transformed genomics research, but they also opened the door to clinical genomics (i.e., molecular diagnostics — see Figure 1). In the clinic, genomic techniques have revolutionized testing across the areas of infectious disease, cancer, and inherited disease by enabling measurement of new analytes, improving analytical performance (e.g., sensitivity) and in some cases providing faster turnaround time compared with traditional testing methods.
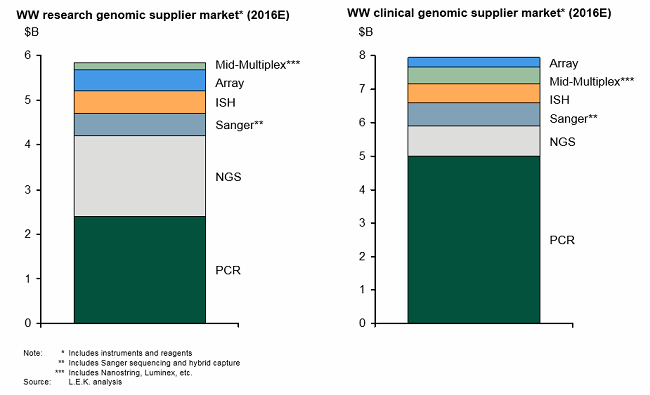
Figure 1: Genomics research leading to clinical genomics
We are only at the beginning of the genomics revolution. What follows are nine key trends and the enabling technologies that will carry the genomics revolution through 2020. (See Figure 2 for how genomics trends have led to the establishment of technology innovators.)
1. Yes, Further Adoption Of NGS
Driven by workflow simplification, continued cost reductions (for both instruments and reagents), and enhanced bioinformatics capabilities, NGS adoption will widen at the high- and low-throughput ends of the research market. There is also broader adoption of NGS-based testing in clinical markets in the areas of oncology, reproductive health, and genetics. Against this backdrop, a number of companies are continuing to innovate (e.g., long-read RNA-sequencing technologies), which will broaden both the research and clinical application sets.
2. Moving To Single-Cell Biology
Currently, most genomic analysis suffers from “average measurements.” In most cases, the sample is lysed, the DNA is extracted and isolated, and analytical measurements are made across different populations (e.g., heterogeneous cells, heterogeneous molecular makeups) in the sample. A number of technology developments may enable researchers to explore biology at the single cell level, thereby eliminating the heterogeneity issue. Those include:
- Sample enrichment: Tools exist (e.g., microfluidics, flow cytometry sorting) that make it possible to partition single cells for subsequent molecular analysis.
- Molecular indexing: Genomic material from individual cells can be uniquely labelled and subsequently pooled for sequencing, enabling researchers to look at gene expression at the individual cell level and compare expression patterns across cell populations, thereby yielding unparalleled insights on cellular heterogeneity.
- Highly sensitive tools: Highly sensitivity genomic technologies (e.g., NGS, digital PCR) permit researchers to explore genomics at the individual cell level, generating new insights into biological processes ranging from normal development to tumor evolution.
3. Emergence Of RNA Biology
An increasing number of publications are highlighting disconcordance between DNA mutations and downstream changes in RNA and/or protein expression. RNA potentially is a more biologically relevant measurement than a surrogate DNA mutation; despite this fact, adoption of RNA analysis in research and clinical markets has failed to keep pace with DNA analysis. Much of this has to do with the difficulties in handling and interrogating RNA. Enabling tools are emerging that can help propel the field of RNA biology — including NGS-based RNA sequencing, which permits researchers to measure the entire transcriptome as well as non-coding RNA, and RNA in situ hybridization (RNA ISH), which enables single-cell analysis of RNA from cell/tissue samples.
4. Emergence Of The “Molecular Stethoscope”
The discovery of cell-free DNA (cfDNA) in circulation has opened the door to what is popularly referred to as the “molecular stethoscope.” The first clinical application of cfDNA was noninvasive pre-natal testing (NIPT), which leverages the ability to detect and interrogate fetal DNA in maternal blood, thus providing a test for fetal chromosomal abnormalities that does not require amniocentesis. Clinical applications of cfDNA extend to cancer (early detection, therapy monitoring, disease recurrence monitoring), pathogen detection for infectious disease, and transplant monitoring. Capturing and analyzing rare cell populations is an extension to the molecular stethoscope concept and takes advantage of single-cell genomics approaches discussed above.
5. Mendelian Genetic Testing
Clinical genomics will vastly improve the entire reproductive health diagnostic paradigm. Genetic testing will begin with carrier screening of mother and father in order to assess more than 250 hereditary diseases. It will extend to in vitro fertilization (IVF), with embryos being examined for inheritance of disease. Additionally, genetic testing will occur in-utero in mothers, analyzing for inherited diseases (e.g., microdeletions). Trio testing, where mother, father, and child are tested, could provide even more powerful information on true inheritance patterns and disease predispositions. Furthermore, population-level genomic data may also yield insights not only into disease, but also into important areas including fertility, resilience, and longevity.
6. Testing Moves Closer To The Patient
Technology advances have automated workflows, decreased instrument footprints, reduced turnaround-time, and simplified test result interpretation. These upgraded capabilities have made it possible for clinical genomic technologies to “decentralize” outside the traditional high-volume central reference laboratories into community hospital labs and physician offices (point-of-care). Continued innovation (e.g., analysis direct from crude samples) will further broaden adoption at the point-of-care, leading to potentially significant improvements in outcomes and patient management economics.
7. Genotype Meets Phenotype
Bioinformatics will unlock genomics across research and clinical markets, but will require integration of siloed datasets. Large-scale efforts such as the 1000 Genomes Project and The Cancer Genome Atlas (TCGA) have generated volumes of genomic data, but these datasets often lack any phenotype or outcomes data. Conversely, EMRs often contain detailed longitudinal patient outcomes data, but here again, data is siloed within a given provider institution. Several efforts (e.g., ASCO CancerLinQ) are underway to link genotype with phenotype in an effort to derive clinical significance from genetic variation.
8. Scaling The Research Experiment
As we discover more genomic biomarkers and unlock genetic diversity, the associated prevalence of these biomarkers will inevitably decrease. To explain the vast genomic differences across populations, researchers will need to conduct experiments at an unprecedented scale. The aforementioned examples (TCGA, 1000 Genomes) are moving into bigger scale studies, but we expect even larger and more coordinated research will be needed (e.g., Million Veteran Program).
9. Clinical Trial Baskets
With the continued unlocking of genetic diversity, we can expect development of more and more targeted therapies aimed at narrow patient populations harboring specific (and increasingly rare) genomic biomarkers. As this unfolds, biopharmaceutical companies will face significant challenges in identifying and enrolling patients for a given drug’s clinical trial. Early coordinated efforts (e.g., Lung Cancer Master Protocol) to address the patient enrollment problem are already underway, but we believe these types of efforts are likely to become more common and make a meaningful impact on drug development.
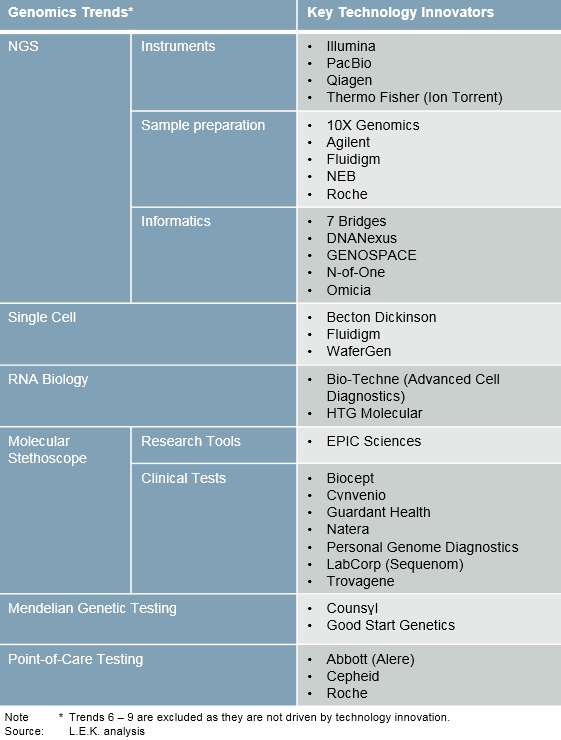
Figure 2: Genomics trends and key innovators
These trends have the potential to create sizable commercial opportunities for in vitro diagnostic (IVD) companies, reference laboratories, life science tools companies and bioinformatics vendors. We encourage these industry participants to develop strategies to help them capitalize on the continued genomics revolution.
About The Authors:
 Alexander Vadas, Ph.D., is a managing director and partner in L.E.K. Consulting’s Biopharmaceuticals & Life Sciences practice. He joined L.E.K. in 2000 and focuses on diagnostics, research tools, and personalized medicine. Within those areas, Vadas has worked with a range of established and emerging clients in the areas of corporate strategy, product strategy, and planning and transaction support. He received both his B.S. and Ph.D. degrees in chemical engineering from the University of California, Los Angeles.
Alexander Vadas, Ph.D., is a managing director and partner in L.E.K. Consulting’s Biopharmaceuticals & Life Sciences practice. He joined L.E.K. in 2000 and focuses on diagnostics, research tools, and personalized medicine. Within those areas, Vadas has worked with a range of established and emerging clients in the areas of corporate strategy, product strategy, and planning and transaction support. He received both his B.S. and Ph.D. degrees in chemical engineering from the University of California, Los Angeles.
 Brian Baranick is a managing director and partner in L.E.K. Consulting's Los Angeles office. He joined L.E.K.’s Biopharma and Life Sciences practice in 2007 and has supported clients across many sectors including biopharmaceuticals, life science tools, diagnostics, and personalized medicine. Baranick also has experience across a broad range of therapeutic area and technology segments. He has advised both established and emerging clients in corporate strategy, product strategy, and planning and transaction support. His thought leadership can be found in several L.E.K. Executive Insights, including Personalized Oncology: A Roadmap and Forging a Path From Companion Diagnostics to Holistic Decision Support. Baranick received his Ph.D. in molecular biology from the University of California, Los Angeles.
Brian Baranick is a managing director and partner in L.E.K. Consulting's Los Angeles office. He joined L.E.K.’s Biopharma and Life Sciences practice in 2007 and has supported clients across many sectors including biopharmaceuticals, life science tools, diagnostics, and personalized medicine. Baranick also has experience across a broad range of therapeutic area and technology segments. He has advised both established and emerging clients in corporate strategy, product strategy, and planning and transaction support. His thought leadership can be found in several L.E.K. Executive Insights, including Personalized Oncology: A Roadmap and Forging a Path From Companion Diagnostics to Holistic Decision Support. Baranick received his Ph.D. in molecular biology from the University of California, Los Angeles.
